If the bits of knowledge from Genetics and Cell Biology are put together things appear to fairly straight forward and conclusive:
- Proteins and enzymes have long chain beaded primary structure made of beads of amino radicals. For each bead location choice could be from one of twenty possible amino radicals which impart them twenty different properties locally.
- Proteins are longer chain molecules which primarily form structures whereas enzymes act and manipulate at molecular level.
- Ultimately it is the specific sequence as well as shape of available “birth cavity” for protein that would determine its tertiary / quarternary structure and its functionality.
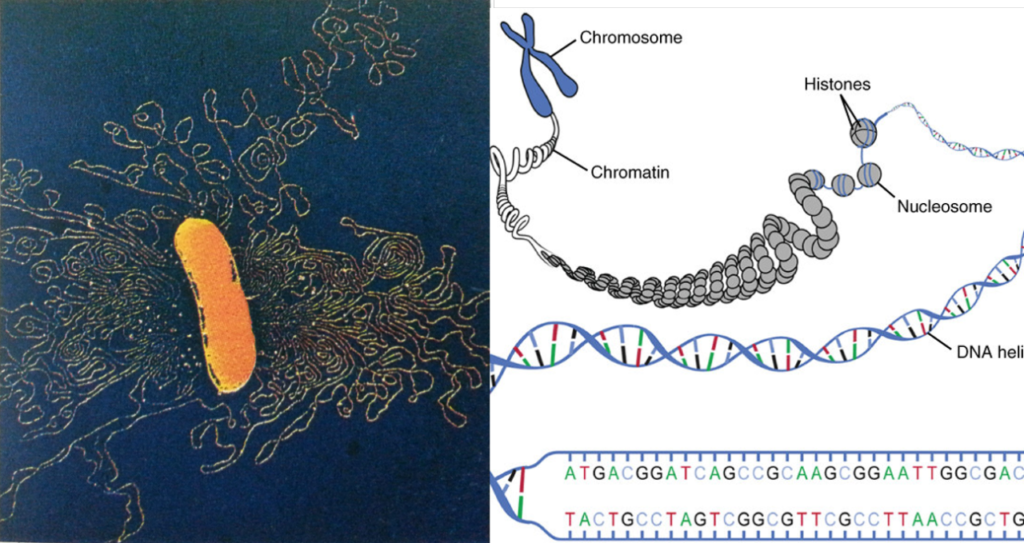
- DNAs have two complimentary “zipped” strands in the form of twisted ladder where the rungs contain A-T pair ( Adenine – Thymine ), T-A ( Thymine – Adenine ), C-G ( Cytosine – Guanine ) or G-C ( Guanine – Cytosine ) when referred from the main code strand.
- Each Amino Radical bead in protein gets coded by three rungs ( bits ? ) on DNA
- Genes are the sections of code which get correlated to specific protein / enzyme
- Complimentary strand is just for closing the zip and stabilization / integrity preservation of the code
- DNA is wound around spools of Histone Balls and they form the Chromatids and Chromosomes.
- Genes have been mapped to specific locations on specific chromosome
- For a protein / enzyme to be produced the corresponding histone bead must be dissolved and the section of twisted ladder has to get “unzipped”
- A slightly different single strand of m-RNA ( messenger ) gets formed picking up the constituents from surrounding solution synthesizing the negative of code where T ( Thymine ) gets replaced by U ( Uracil ) so that it separates from double helix easily.
- The “transcripted” m-RNA extrudes its way until it reaches r-RNA ( ribosomal RNA ) which has a vise and ratchet type of mechanism to pull the m-RNA strand and feed it in increments of three rungs …the triplet that is held in it gets translated into corresponding amino radical bead which gets incorporated the protein that gets extruded at the other side and protein strand gets delivered at specific location depending on location of r-RNA..
- Pick the matching bead from solution gets done by t-RNA ( transfer RNA ) which eventually controls the “translation” and defines the correspondence of genetic code uniquely
Well, so far so good BUT there are many inadequately explained mysteries:
- What causes appropriate histone bead to melt and reappear after the job is done?
- What causes “unzipping” of DNA strands for the protein to come OR for protein “on-demand”? What triggers its “re-zipping”
- What dictates which genes shall be expressed / translated selectively for a kind of cell?
- How are those “bookmarks” moved for oncogenesis of a different type of cell?
- How do m-RNAs find the right r-RNAs at their location OR is the complete assembly including r-RNA and the related delivery and navigational cavities are all formed in one go.
- What controls how many “transcripts” and proteins get produced from a gene? Is there an inbuilt “counter” or is it a feedback based closure?
- If the strand gets shifted by one / two rung the resulting code would be totally different …how the “framing” error gets prevented while transcripting and translating?
- What prevents the complimentary strand to be transcripted and translated? It would be entirely different end product.
- There is certainly a logical need for “Reverse Transcription” and “Reverse Translation” whereby an existing protein could get encoded and substituted on mapped gene but there is no mention of m-RNA, t-RNA and r-RNA being reversible.
For most of above genetic “who-done-it?” the generic answer proposed is “nuclear enzymes” BUT what controls them and how exactly do they work at molecular dimension level robustly in a semi fluid media remains a grey area.
AVINASH KHARE